The AACR Annual Meeting went virtual for the second year in a row in 2021. The cancer research community has done an admirable job of adapting to the new format – AACR brought together leaders in the field to share new research and discuss the future of cancer treatment.
While we miss meeting our users in-person (and grabbing hundreds of pens in the exhibitor hall), there is one aspect of the virtual format we enjoy: on-demand presentations (and watching them in our PJs). Spatial biology was featured in several talks at the meeting, and we were able to watch our favorites on repeat. Here’s a roundup of our top three.
We’ll highlight some posters from the conference in part two of this series – stay tuned!
Novel analytical framework reveals cellular neighborhoods in the tumor microenvironment
Pathology from the atomic scale on up – Dr. Garry Nolan

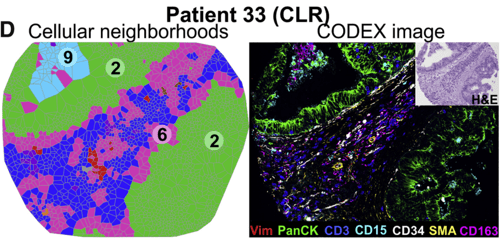
Dr. Garry Nolan of Stanford University, a well-recognized thought leader in the field of immune biology, gave a plenary talk during the conference and introduced a novel analytical framework, developed by his lab, to uncover distinct “cellular neighborhoods” in the tumor microenvironment. These neighborhoods are each uniquely composed of certain immune and cancer cell types.
This seminal spatial biology algorithm was developed with data generated solely on the CODEX multiplex imaging system and the results were published in Cell in September 2020. Dr. Nolan demonstrated how the organization and interactions of these neighborhoods predicted patient outcomes in colorectal cancer and cutaneous T cell lymphoma, underscoring the importance of spatial context in developing prognostic biomarkers.
Representative Voronoi diagrams of cellular neighborhoods (CNs) in colorectal cancer (left) and corresponding seven-color image (right). Image: Schürch et al (2020) / CC BY 4.0.
Validating multiplex immunofluorescence for clinical research applications
Multiplex immunofluorescence assay development — Dr. Janis Taube

Dr. Janis Taube of Johns Hopkins University School of Medicine, a pioneer in mIF applications, discussed best practices for mIF assay development and validation as part of a session on future directions for immunotherapy diagnostics. She also shared findings from the MITRE study, the first ever multi-institutional endeavor to validate an mIF workflow for immuno-oncology – developed on the Phenoptics solution (Opal).
“The next generation of tissue-based biomarkers will likely include the identification and quantification of multiple cell types and their spatial interactions,” said Dr. Taube. “Multiplex technologies are especially adept at defining these relationships.”
To ensure high-quality data from her mIF assays, Dr. Taube described her team’s analysis pipeline, which includes staining, image acquisition, image processing, phenotyping, normalizing batch effects, and data handling. Her guiding principle when developing and validating mIF assays is to confirm that each marker in the assay stains at levels comparable to chromogenic IHC, the current gold standard, with the additional goal of robust assessment of marker expression intensity in situ.
As part of the MITRE study, Dr. Taube collaborated with industry and academic groups to perform reproducibility assessments of a six-plex Phenoptics mIF assay on tonsil and tumor tissue microarrays (TMAs) across six institutions, finding high inter- and intra-site reproducibility. The results from the study speak to the robustness of the Phenoptics mIF solution and its readiness for clinical applications.
Spatial biomarker signatures predict treatment response in breast cancer
The neoadjuvant platform trials as an engine for optimization for immuno and other therapy – Dr. Laura J Esserman

Dr. Laura Esserman, from the University of California San Francisco (UCSF) is the principal investigator of the i-SPY Trials. The i-SPY Trials are very biomarker-rich, noted Dr. Esserman. They were built around the idea that breast cancer is heterogenous, thus requiring many different types of biomarkers to measure that heterogeneity.
In her talk, Dr. Esserman demonstrated the use of mIF-based biomarker panels, developed with the Phenoptics platform, to map the heterogeneity of the tumor microenvironment. Dr. Esserman’s team obtained spectrally unmixed tissue images to perform cell segmentation, automated phenotyping and cell counts, and spatial analyses.
The team sought to identify which immune cell populations were associated with pathologic complete response (pCR) in breast cancer. They measured the spatial proximity between different cell types, such as T cells and tumor cells, identifying spatial biomarker signatures that correlate with pCR.
Interested in learning how spatial biology is enabling the Cancer Immunome Project? Register for our upcoming webinar.