This year’s AACR Annual Meeting was an entirely different experience. Held online and split into two events, the meeting still managed to bring together the cancer research community – the first half of the conference in April drew over 60,000 attendees alone.
There were multiple posters at AACR demonstrating the applications of CODEX & Phenoptics in cancer research. We presented a few with exciting new data at the second part of the conference in June. Read the summary of the findings below, or click the thumbnails to download the posters.
We’ve also included a poster from the lab of Dr. Garry Nolan at Stanford University, which uses CODEX data to identify predictive biomarkers for immunotherapy.
Developing and Validating a High-throughput 9-color Immunofluorescence Assay for Translational Studies in Immuno-oncology Research
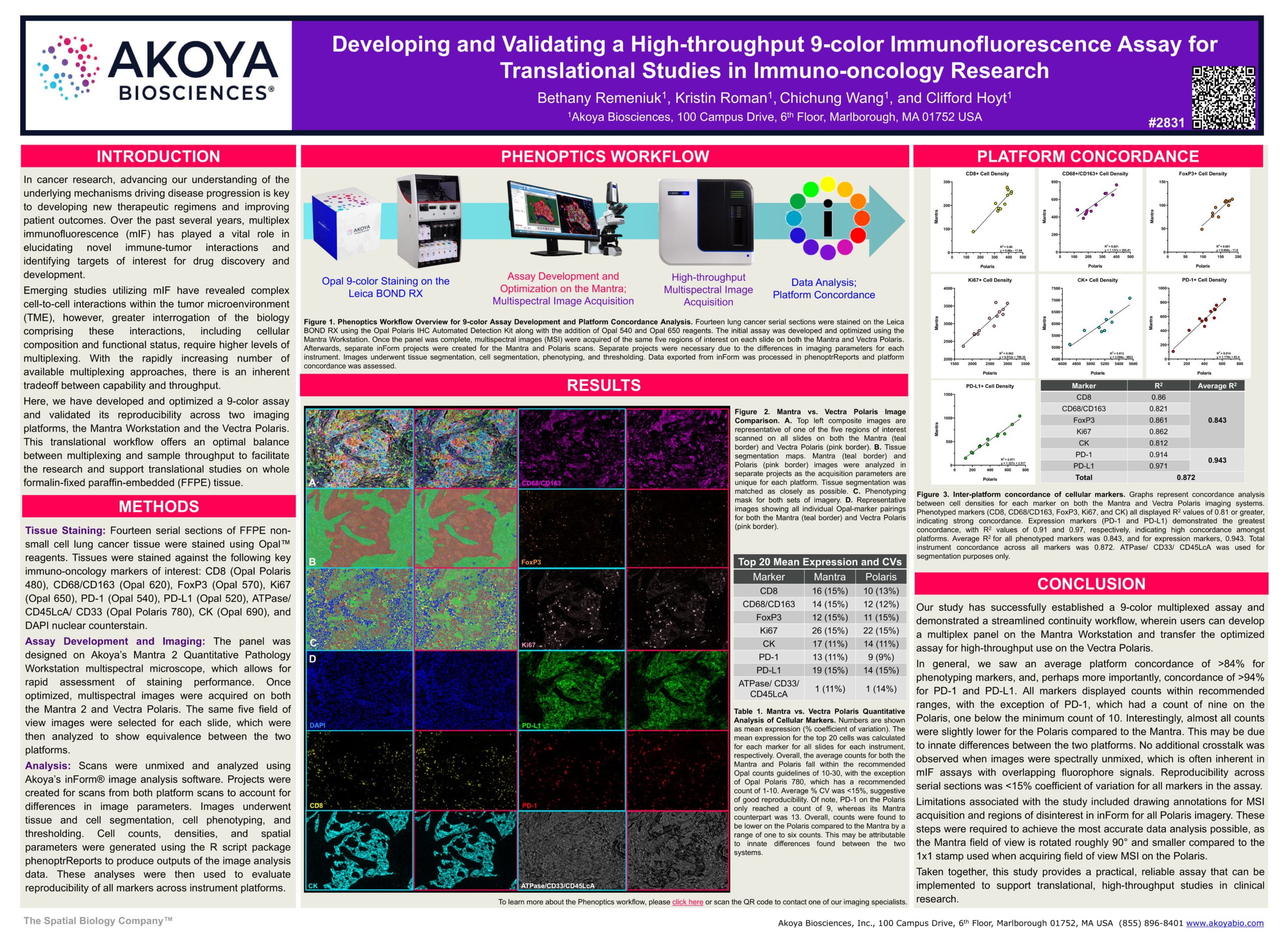
This poster describes a translational workflow offering an optimal balance between multiplexing and sample throughput to facilitate research and support translational studies on whole formalin-fixed, paraffin-embedded (FFPE) tissue. We developed and optimized a 9-color assay and validated its reproducibility across two imaging platforms: the Mantra 2 Quantitative Pathology Workstation and the Vectra Polaris.
Our team designed the panel using the Mantra, which allows for rapid assessment of staining performance. After optimization, images were acquired on both the Mantra and Vectra Polaris. We selected the same five field-of-view images for each slide and analyzed them to show equivalence between the platforms.
The mean expression (% coefficient of variation) for the top 20 cells was calculated for each marker and for all slides on each instrument. The average counts for both the Mantra and Polaris fell within the recommended Opal counts guidelines, and the average % CV was <15%, which suggests good reproducibility. We observed an average platform concordance of >84% for phenotypic markers and a concordance of >94% for PD-1 and PD-L1.
This study demonstrates a streamlined workflow, wherein users can develop a multiplex panel on the Mantra and then transfer the optimized assay for high-throughput use on the Vectra Polaris.
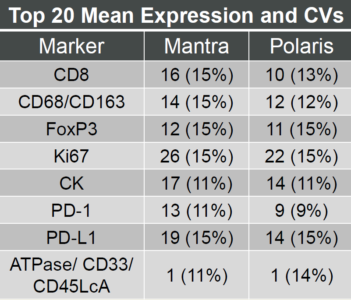
Analysis of FFPE human tumor tissues using CODEX with signal amplification
We recently expanded our CODEX workflow to amplify the fluorescent signal intensities of low expressing biomarkers by combining Tyramide Signal Amplification (TSA) with CODEX, also known as CODEX® Amp. In this poster, we present the analysis of CODEX data for a panel of over 20 biomarkers on human FFPE tissue, including signal amplification of PD-L1, PD-1 and FOXP3.
The CODEX Amp workflow involves the following steps: Tissues are stained offline in a single step according to standard CODEX protocol. After iterative cycles of labelling, imaging, and removing via our standard CODEX workflow, up to three cycles of HRP oligonucleotides, catalysis, deposition of a single CODEX Amp dye per cycle, and removal of HRP oligonucleotides can be performed and imaged with our standard microscope filters (Cy3, Cy5, AF750).
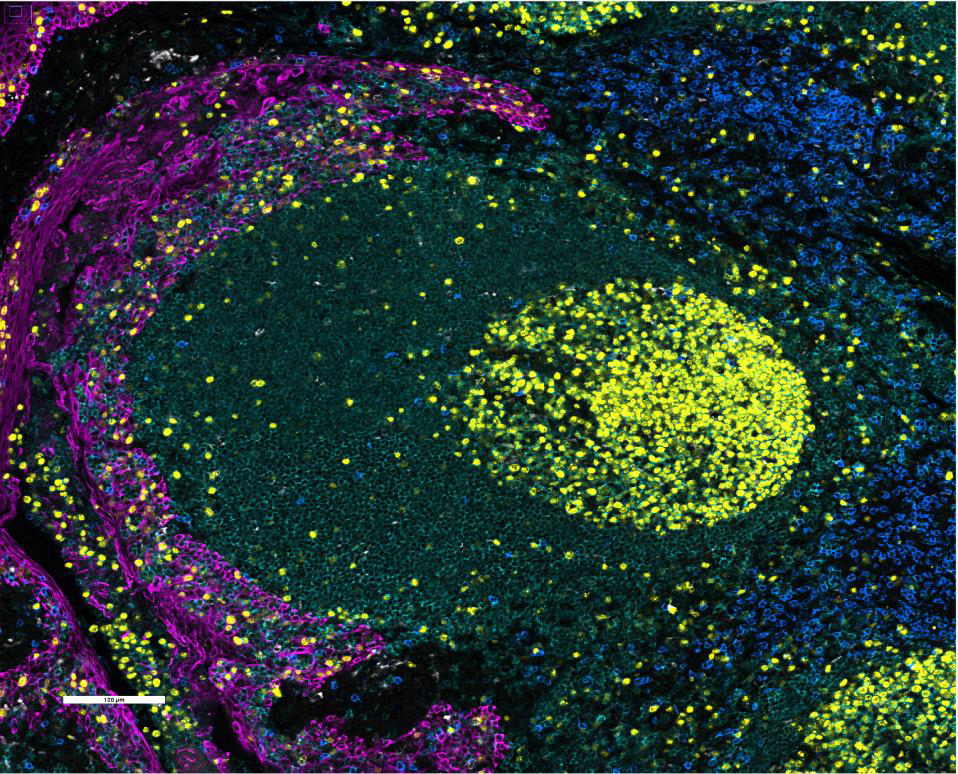
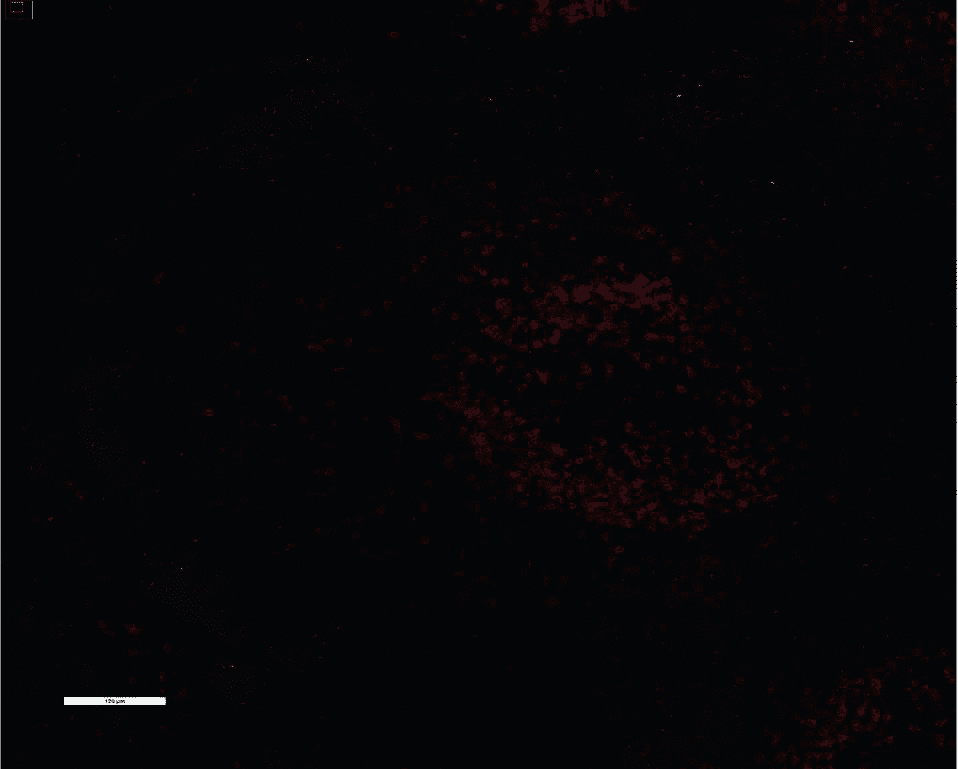
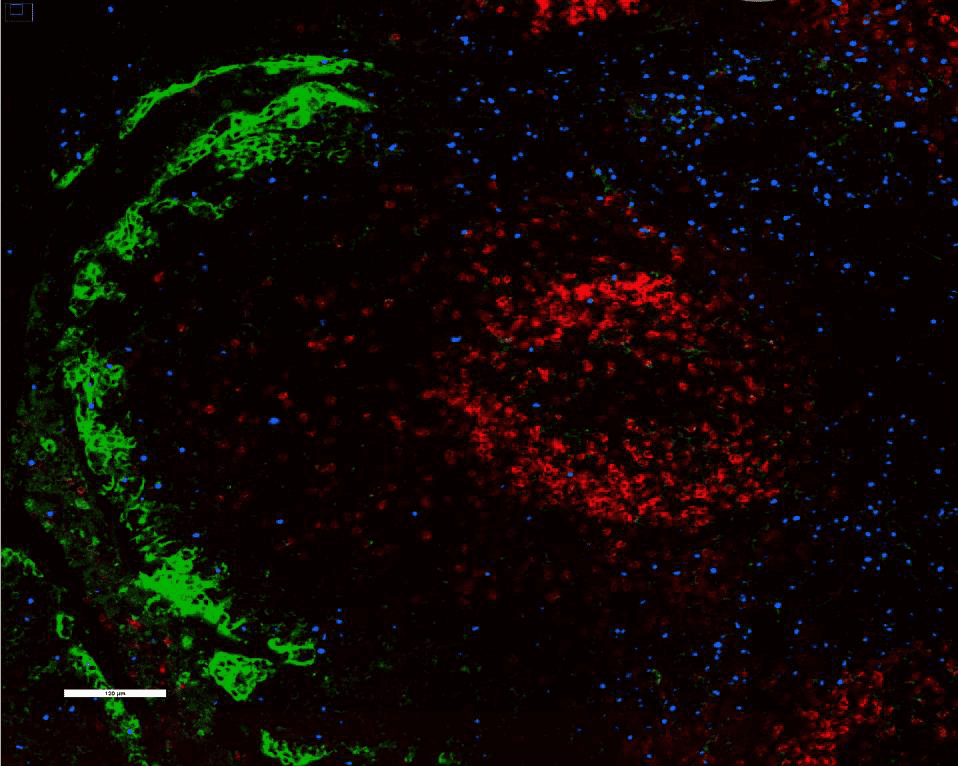
We stained and imaged standard CODEX markers (left) CD20, CD68, PCK, CD8, and Ki67 and key IO markers (middle) PD-L1, PD-1, and FoxP3 on FFPE human tonsil with the standard CODEX workflow. PD-L1, PD-1, and FoxP3 antibodies were then amplified using the CODEX Amp workflow and imaged (right). A one-way ANOVA test and post-hoc analysis indicated statistically significant differences between each group.
Highly multiplexed analysis of FFPE breast tissues using the CODEX® technology

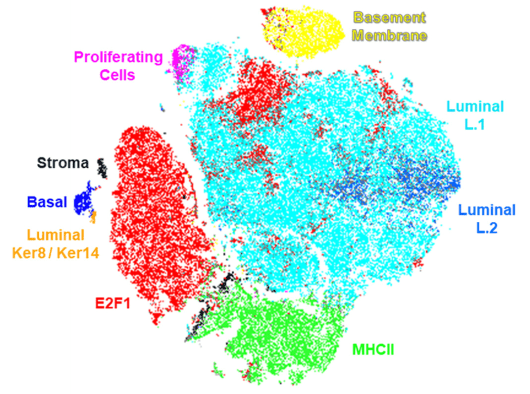
This poster demonstrates the development of a 36-marker breast cancer-specific CODEX panel, which was designed to identify different subpopulations of cells in FFPE breast cancer tissue. We generated data on the CODEX platform for the different biomarkers using FFPE Human breast cancer tissues at different stages as well as normal FFPE Human breast tissue. This panel enabled us to detect cancer cells as well as non-malignant cells, which are also part of the tumor microenvironment, to enable a better understanding of communication between the cells.
We also tested CODEX’s software capabilities — a deep phenotyping analysis was conducted for Ductal Adenocarcinoma (Stage IIB) to demonstrate its powerful functionality. Each biomarker signal was quantified on a per-cell basis and clusters were defined based on these signals. Phenotypic relationships were analyzed using t-SNE plots, which allow the flexibility to toggle between the plot and the original morphological data and simultaneously examine spatial context and associations between different types of cells. We were able to identify ten different subpopulations of the cells including a rare Luminal Ker8+/Ker14+.
For more information about this panel, watch this on-demand webinar.
Cellular neighborhoods predict pembrolizumab response in cutaneous T cell lymphomas
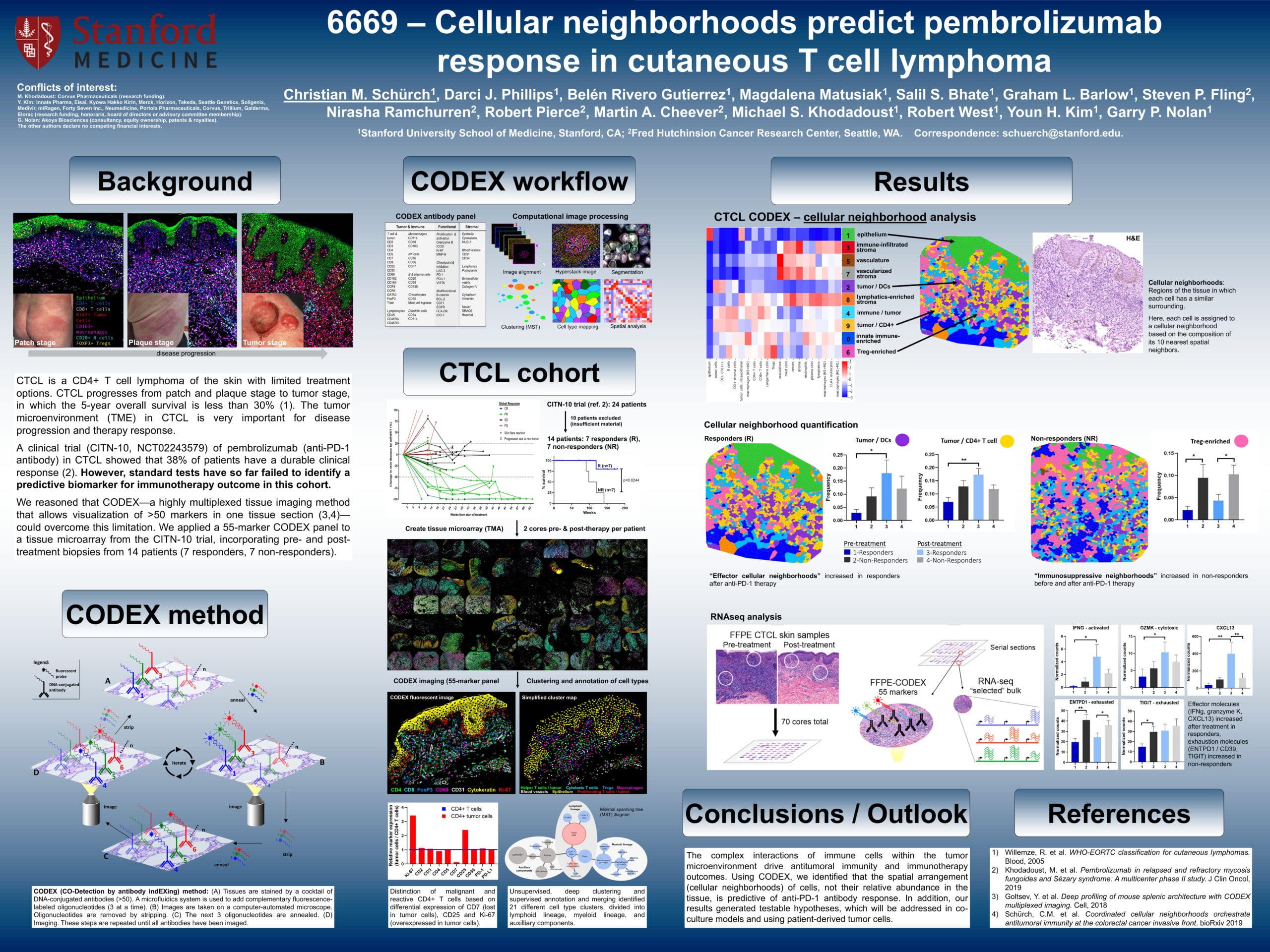
Cutaneous T cell lymphoma (CTCL) is a rare cancer type which affects T cells, causing them to attack the skin and result in skin lesions. CTCL eventually progresses from the patch and plaque stages to the tumor stage.
In a clinical trial, the anti-PD-1 immunotherapy, pembrolizumab, was shown to elicit a durable clinical response in 38% of patients with CTCL. However, as noted in the poster, current testing has not yet identified a biomarkers to predict immunotherapy outcomes for this cohort.
Schürch et. al. used a 55-marker CODEX panel on a tissue microarray from the clinical trial. They performed identified cellular neighborhoods — each cell was assigned to a neighborhood based on the composition of its 10 nearest spatial neighbors. They observed that “effector cellular neighborhoods” increased in responders following anti-PD-1 treatment, while “immunosuppressive neighborhoods” increased in non-responders before and after treatment.
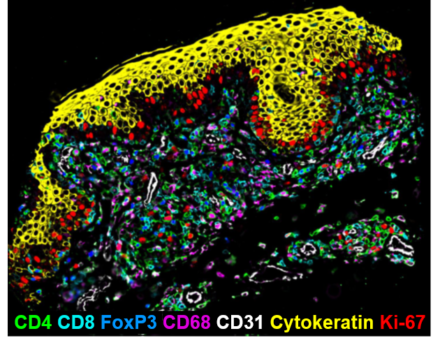
These results indicate that the spatial arrangement of cells, rather than their relative abundance, is predictive of anti-PD-1 immunotherapy response. The researchers plan to follow up this study by testing further hypotheses in co-culture models and patient-derived tumor cells.
Curious about any of the data presented here? We’re happy to answer your questions.





